Pneumonia is an infection that inflames the air sacs in one or both lungs. The air sacs may fill with fluid or pus (purulent material), causing cough with phlegm or pus, fever, chills, and difficulty breathing. A variety of organisms, including bacteria, viruses and fungi, can cause pneumonia. An early detection and treatment to prevent progression might be crucial due to its fatality rate, which is 5 to 10 percent for hospitalized patients.
This study aims to provide a classification model trained with the X-Ray images provided in the pneumonia detection challenge in Kaggle. Further information regarding the dataset can be found in the ChestX-ray8 paper.
A Colab-friendly version of this notebook is available at GitHub, which can be directly run on Google Colab with a valid Kaggle credential file.
Importing Libraries
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import pydicom # To read dicom files
# Modeling --------------------------
import torch
import torchvision
from torchvision import transforms # For data augmentation & normalization
import torchmetrics # Easy metric computation
import pytorch_lightning as pl
from pytorch_lightning.callbacks import ModelCheckpoint
from pytorch_lightning.loggers import TensorBoardLogger
# -----------------------------------
from pathlib import Path # For file path handling
import cv2 # For image resizing
from tqdm.notebook import tqdm # For progress bar
import os
Preprocessing and Loading the Data
1
2
3
4
# Reading the data labels
labels = pd.read_csv("stage_2_train_labels.csv")
labels.head(10)
| patientId | x | y | width | height | Target | |
|---|---|---|---|---|---|---|
| 0 | 0004cfab-14fd-4e49-80ba-63a80b6bddd6 | NaN | NaN | NaN | NaN | 0 |
| 1 | 00313ee0-9eaa-42f4-b0ab-c148ed3241cd | NaN | NaN | NaN | NaN | 0 |
| 2 | 00322d4d-1c29-4943-afc9-b6754be640eb | NaN | NaN | NaN | NaN | 0 |
| 3 | 003d8fa0-6bf1-40ed-b54c-ac657f8495c5 | NaN | NaN | NaN | NaN | 0 |
| 4 | 00436515-870c-4b36-a041-de91049b9ab4 | 264.0 | 152.0 | 213.0 | 379.0 | 1 |
| 5 | 00436515-870c-4b36-a041-de91049b9ab4 | 562.0 | 152.0 | 256.0 | 453.0 | 1 |
| 6 | 00569f44-917d-4c86-a842-81832af98c30 | NaN | NaN | NaN | NaN | 0 |
| 7 | 006cec2e-6ce2-4549-bffa-eadfcd1e9970 | NaN | NaN | NaN | NaN | 0 |
| 8 | 00704310-78a8-4b38-8475-49f4573b2dbb | 323.0 | 577.0 | 160.0 | 104.0 | 1 |
| 9 | 00704310-78a8-4b38-8475-49f4573b2dbb | 695.0 | 575.0 | 162.0 | 137.0 | 1 |
The data includes 6 columns including patient ID, target variable, and information about the location of the pneumonia if it exists. Notice that there are multiple entries for some patients since pneumonia can be located at more than one segment of an X-Ray image. A subset of the dataset such that each patient has only one record is sufficient in this case since the aim of this study is not detection.
1
2
3
# Dropping the duplicate rows based on patient ID
labels = labels.drop_duplicates("patientId")
1
2
3
4
# Defining import and export paths
ROOT_PATH = Path("stage_2_train_images/")
SAVE_PATH = Path("Processed/")
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
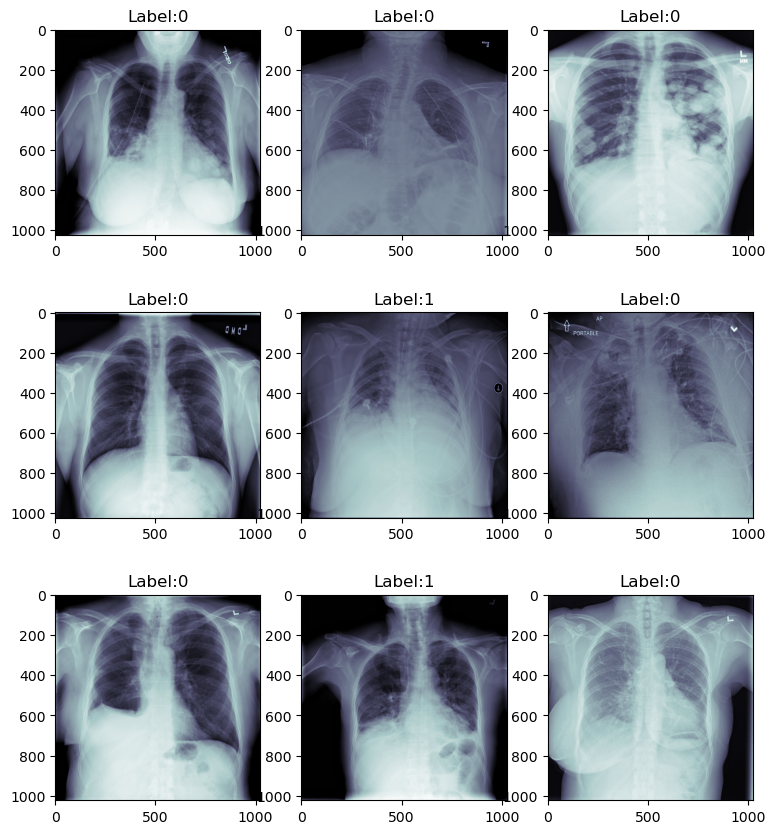
# Writing a nested loop to read and visualize a sample from the dataset
fig, axis = plt.subplots(3, 3, figsize = (9, 9))
plt.subplots_adjust(top=1)
c = 0
for i in range(3):
for j in range(3):
# Load image
patient_id = labels.patientId.iloc[c]
dcm_path = ROOT_PATH/patient_id
dcm_path = dcm_path.with_suffix(".dcm")
dcm = pydicom.read_file(dcm_path).pixel_array
# Print image
label = labels["Target"].iloc[c]
axis[i][j].imshow(dcm, cmap="bone")
axis[i][j].set_title(label)
c += 1
Figure 1. Images Before Augmentation
1
2
3
4
len_train = len(os.listdir(ROOT_PATH))
len_valid = len(os.listdir("stage_2_test_images/"))
print(len_train)
print(len_valid)
1
2
26684
3000
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
# Resizing the images for an easy training
# Standardazing the pixel values by dividing by 255
# Using tqdm to track the loading progress
sums, sums_squared = 0, 0
for c, patient_id in enumerate(tqdm(labels.patientId)):
# Load image
patient_id = labels.patientId.iloc[c]
dcm_path = ROOT_PATH/patient_id
dcm_path = dcm_path.with_suffix(".dcm")
dcm = pydicom.read_file(dcm_path).pixel_array / 255
dcm_array = cv2.resize(dcm, (224, 224)).astype(np.float16)
label = labels.Target.iloc[c]
# Splitting the data into training and test
train_or_test = "train" if c < 24000 else "test"
current_save_path = SAVE_PATH/train_or_test/str(label)
current_save_path.mkdir(parents=True, exist_ok=True)
np.save(current_save_path/patient_id, dcm_array)
normalizer = 224 * 224
if train_or_test == "train":
sums += np.sum(dcm_array) / normalizer
sums_squared += (np.power(dcm_array, 2).sum()) / normalizer
1
0%| | 0/26684 [00:00<?, ?it/s]
1
2
3
4
5
# Defining and checking the mean and the standard deviation
mean = sums / len_train
stdev = np.sqrt(sums_squared / len_train - (mean**2))
print(f"Mean:\t\t\t {mean} \nStandard Deviation:\t {stdev}")
1
2
Mean: 0.44106992823128194
Standard Deviation: 0.27758244159100576
1
2
3
4
# Function to load data
def load_file(path):
return(np.load(path).astype(np.float32))
The following cells perform random augmentations on the dataset such as crops, rotations etc. to make the model more powerful in assessing low-quality images.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
# Data Augmentation Settings
train_transforms = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean, stdev),
transforms.RandomAffine(degrees = (-5, 5),
translate = (0, 0.05),
scale = (0.9, 1.1)),
transforms.RandomResizedCrop((224, 224),
scale = (0.35, 1))
])
test_transforms = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean, stdev),
])
1
2
3
4
5
6
7
8
9
10
11
# Defining the train and test data
train = torchvision.datasets.DatasetFolder("Processed/train/",
loader = load_file,
extensions = "npy",
transform = train_transforms)
test = torchvision.datasets.DatasetFolder("Processed/test/",
loader = load_file,
extensions = "npy",
transform = test_transforms)
1
2
3
4
5
6
7
8
9
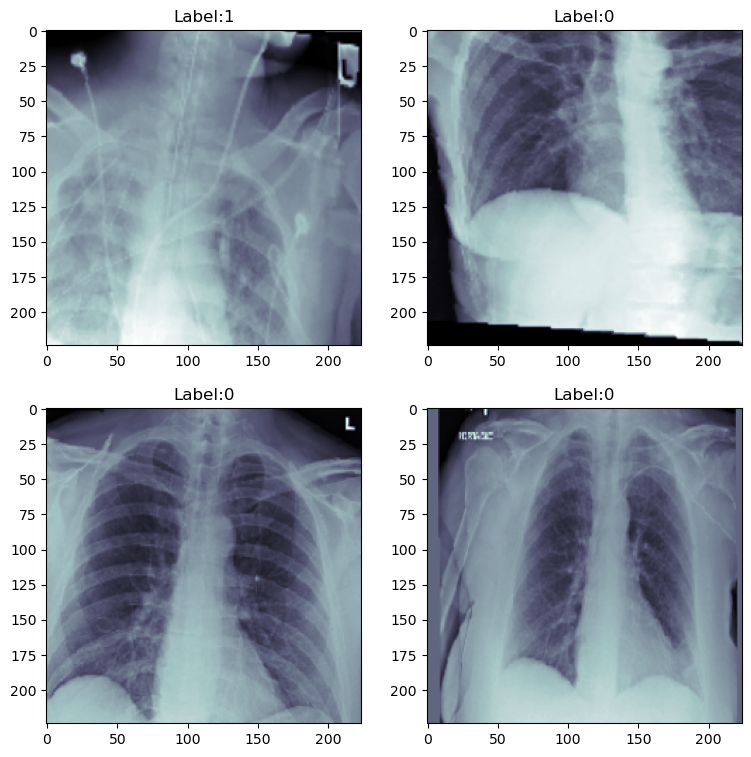
# Viewing a random sample of 4
fig, axis = plt.subplots(2, 2, figsize = (9, 9))
for i in range(2):
for j in range(2):
random_index = np.random.randint(0, 24000)
x_ray, label = train[random_index]
axis[i][j].imshow(x_ray[0], cmap="bone")
axis[i][j].set_title(f"Label:{label}")
Figure 2. Images After Augmentation
The effect of augmentation can be seen clearly.
1
2
3
4
5
6
7
batch_size = 64
num_workers = 2
train_loader = torch.utils.data.DataLoader(train, batch_size = batch_size, num_workers = num_workers, shuffle = True)
test_loader = torch.utils.data.DataLoader(test, batch_size = batch_size, num_workers = num_workers, shuffle = False)
print(f"# of train images: \t{len(train)} \n# of test images: \t{len(test)}")
1
2
# of train images: 24000
# of test images: 2684
1
2
3
# Checking the number of images with and without pneumonia in the train set
np.unique(train.targets, return_counts = True)
1
(array([0, 1]), array([18593, 5407]))
Since the data is imbalanced and the number of images without pneumonia are almost 3 times higher than images with pneumonia, a weighted loss of 3 can be used in the model, which means that the model will assign a higher penalty for the misclassification of the negative class.
Modeling
We are going to use ResNet-18, which is an 18 layers deep convolutional neural network, along with the optimizer Adam. The architecture of ResNet-18 can be checked with the following code:
1
torchvision.models.resnet18()
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
ResNet(
(conv1): Conv2d(3, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)
(bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(maxpool): MaxPool2d(kernel_size=3, stride=2, padding=1, dilation=1, ceil_mode=False)
(layer1): Sequential(
(0): BasicBlock(
(conv1): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
(1): BasicBlock(
(conv1): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(layer2): Sequential(
(0): BasicBlock(
(conv1): Conv2d(64, 128, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(downsample): Sequential(
(0): Conv2d(64, 128, kernel_size=(1, 1), stride=(2, 2), bias=False)
(1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): BasicBlock(
(conv1): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(128, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(layer3): Sequential(
(0): BasicBlock(
(conv1): Conv2d(128, 256, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(downsample): Sequential(
(0): Conv2d(128, 256, kernel_size=(1, 1), stride=(2, 2), bias=False)
(1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): BasicBlock(
(conv1): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(256, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(layer4): Sequential(
(0): BasicBlock(
(conv1): Conv2d(256, 512, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(downsample): Sequential(
(0): Conv2d(256, 512, kernel_size=(1, 1), stride=(2, 2), bias=False)
(1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): BasicBlock(
(conv1): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn1): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(relu): ReLU(inplace=True)
(conv2): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): BatchNorm2d(512, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(avgpool): AdaptiveAvgPool2d(output_size=(1, 1))
(fc): Linear(in_features=512, out_features=1000, bias=True)
)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
class PneumoniaClassifier(pl.LightningModule):
def __init__(self):
super().__init__()
self.model = torchvision.models.resnet18()
# modifying the input channels of the first convolutional layer (conv1) from 3 to 1
self.model.conv1 = torch.nn.Conv2d(1, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)
# modifying the out_features of the last fully connected layer (fc) from 1000 to 1
self.model.fc = torch.nn.Linear(in_features=512, out_features=1, bias=True)
self.optimizer = torch.optim.Adam(self.model.parameters(), lr=1e-4)
# Adding the weigted loss to overcome the imbalance in the dataset
self.loss_func = torch.nn.BCEWithLogitsLoss(pos_weight=torch.tensor([3]))
# Tracking the train and validation accuracy
self.train_acc = torchmetrics.Accuracy(task="binary")
self.valid_acc = torchmetrics.Accuracy(task="binary")
# ---
self.training_step_outputs = []
self.validation_step_outputs = []
def forward(self, data):
# Computes the output of ResNet-18 and returns the prediction
pred = self.model(data)
return(pred)
def training_step(self, batch, batch_idx):
# PyTorch lightning optimizes according to the value returned by this function
# Calculating the loss
x_ray, label = batch
label = label.float()
pred = self(x_ray)[:,0]
loss = self.loss_func(pred, label)
# ---
self.training_step_outputs.append(loss)
# Recording accuracy
self.log("Train Loss", loss)
self.log("Step Train Acc", self.train_acc(torch.sigmoid(pred), label.int())) # Converted to probability w/Sigmoid
return(loss)
def on_train_epoch_end(self):
epoch_average = torch.stack(self.training_step_outputs).mean()
self.log("training_epoch_average", epoch_average)
self.training_step_outputs.clear() # free memory
def validation_step(self, batch, batch_idx):
x_ray, label = batch
label = label.float()
pred = self(x_ray)[:,0]
loss = self.loss_func(pred, label)
# ---
self.validation_step_outputs.append(loss)
self.log("Validation Loss", loss)
self.log("Step Validation Acc", self.valid_acc(torch.sigmoid(pred), label.int()))
return(loss)
def on_validation_epoch_end(self):
epoch_average = torch.stack(self.validation_step_outputs).mean()
self.log("validation_epoch_average", epoch_average)
self.validation_step_outputs.clear() # free memory
def configure_optimizers(self):
return([self.optimizer])
1
model = PneumoniaClassifier()
1
2
available_gpus = [torch.cuda.device(i) for i in range(torch.cuda.device_count())]
available_gpus
1
[<torch.cuda.device at 0x7f1ec6def6a0>]
1
2
3
4
5
6
7
8
9
10
11
12
13
# Model checkpoint: Save top 10 checkpoints based on the highest validation accuracy
checkpoint_callback = ModelCheckpoint(monitor = 'Step Validation Acc',
save_top_k = 10,
mode = 'max')
# Creating the trainer
trainer = pl.Trainer(accelerator = "gpu",
logger = TensorBoardLogger(save_dir= "./logs"),
log_every_n_steps = 1,
callbacks = checkpoint_callback,
max_epochs = 40)
1
2
3
4
INFO:pytorch_lightning.utilities.rank_zero:GPU available: True (cuda), used: True
INFO:pytorch_lightning.utilities.rank_zero:TPU available: False, using: 0 TPU cores
INFO:pytorch_lightning.utilities.rank_zero:IPU available: False, using: 0 IPUs
INFO:pytorch_lightning.utilities.rank_zero:HPU available: False, using: 0 HPUs
1
trainer.fit(model, train_loader, test_loader)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
INFO:pytorch_lightning.accelerators.cuda:LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
INFO:pytorch_lightning.callbacks.model_summary:
| Name | Type | Params
------------------------------------------------
0 | model | ResNet | 11.2 M
1 | loss_func | BCEWithLogitsLoss | 0
2 | train_acc | BinaryAccuracy | 0
3 | valid_acc | BinaryAccuracy | 0
------------------------------------------------
11.2 M Trainable params
0 Non-trainable params
11.2 M Total params
44.683 Total estimated model params size (MB)
INFO:pytorch_lightning.utilities.rank_zero:`Trainer.fit` stopped: `max_epochs=40` reached.
Validation
1
2
3
4
5
# Set device to cuda if available
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
model.eval()
model.to(device)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
# Calculating predictions
preds = []
labels = []
with torch.no_grad():
for data, label in tqdm(test):
data = data.to(device).float().unsqueeze(0)
pred = torch.sigmoid(model(data)[0].cpu())
preds.append(pred)
labels.append(label)
preds = torch.tensor(preds)
labels = torch.tensor(labels).int()
1
2
3
4
5
6
7
8
9
10
11
# Checking the metrics
acc = torchmetrics.Accuracy(task="binary")(preds, labels)
precision = torchmetrics.Precision(task="binary")(preds, labels)
recall = torchmetrics.Recall(task="binary")(preds, labels)
cm = torchmetrics.ConfusionMatrix(task="binary")(preds, labels)
print(f"Accuracy:\t\t{acc}")
print(f"Precision:\t\t{precision}")
print(f"Recall:\t\t\t{recall}")
print(f"Confusion Matrix:\n {cm}")
1
2
3
4
5
6
Accuracy: 0.8036512732505798
Precision: 0.5432372689247131
Recall: 0.8099173307418823
Confusion Matrix:
tensor([[1667, 412],
[ 115, 490]])
| Real Label | |||
|---|---|---|---|
| Positive | Negative | ||
| Predicted Label | Positive | 490 | 412 |
| Negative | 115 | 1667 | |
Table 1. Confusion Matrix
High recall points out that the model rarely misses the cases with pneumonia, yet the precision score is not that good and points out the high number of false positives. In this context, the model’s performance can be considered good since missing a pneumonia case is worse than predicting a false positive.
For lower maximum number of epochs, the model would yield a better accuracy and worse recall, yet a better recall and a sufficient accuracy is obviously better. Therefore, it can be concluded that setting the penalty weight to 3 contributed well, as the model gets closer to the optimum with higher epochs.
Deep Learning with PyTorch for Medical Image Analysis
GitHub Repository